New & Noteworthy
Updates to legacy gene names
November 05, 2021
SGD has long been the keeper of the official Saccharomyces cerevisiae gene nomenclature. Robert Mortimer handed over this responsibility to SGD in 1993 after maintaining the yeast genetic map and gene nomenclature for 30 years.
The accepted format for gene names in S. cerevisiae comprises three uppercase letters followed by a number. The letters typically signify a phrase (referred to as the “Name Description” in SGD) that provides information about a function, mutant phenotype, or process related to that gene, for example “ADE” for “ADEnine biosynthesis” or “CDC” for “Cell Division Cycle”. Gene names for many types of chromosomal features follow this basic format regardless of the type of feature named, whether an ORF, a tRNA, another type of non-coding RNA, an ARS, or a genetic locus. Some S. cerevisiae gene names that pre-date the current nomenclature standards do not conform to this format, such as MRLP38, RPL1A, and OM45.
A few historical gene names predate both the nomenclature standards and the database, and were less computer-friendly than more recent gene names, due to the presence of punctuation. SGD recently updated these gene names to be consistent with current standards and to be more software-friendly by removing punctuation. The old names for these four genes have been retained as aliases.
| ORF | Old gene name | New gene name |
|---|---|---|
| YGL234W | ADE5,7 | ADE57 |
| YER069W | ARG5,6 | ARG56 |
| YBR208C | DUR1,2 | DUR12 |
| YIL154C | IMP2′ | IMP21 |
Categories: Announcements, Data updates
Tags: gene nomenclature
Reference Genome Annotation Update R64.3
August 03, 2021
The S. cerevisiae strain S288C reference genome annotation was updated in its first major update since 2014. The new genome annotation is release R64.3, which released on April 21, 2021. Note that the underlying sequence of 16 assembled nuclear chromosomes, plus the mitochondrial genome, remained unchanged in annotation release R64.3.1 (relative to genome sequence release R64.2.1).
This annotation update included:
- 7 new ORFs: OTO1/YGR227C-A, YHR052C-B, YHR054C-B, YJR107C-A, YKL104W-A, YLR379W-A, YMR008C-A
- 5 new ncRNAs: GAL10-ncRNA, TBRT/XUT_2F-154, SUT169, PHO84 lncRNA, GAL4 lncRNA
- 2 new uORFs for ROK1/YGL171W
- 1 new recombination enhancer: RE
- 1 new LTR: YELWdelta27
- 3 ORFs with shifted translation starts: HPA3/YEL066W, YJR012C, LTO1/YNL260C
- 1 ORF with shifted translation stop plus new intron: LDO45/YMR147W
- Changed feature_type (and SO_term) for non-transcribed spacers: NTS1-2, NTS2-1, NTS2-2
- New systematic nomenclature system for entire annotated complement of ncRNAs
R64.3 Annotation update details
| Chr | Feature | Description of change | Reference |
|---|---|---|---|
| II | YNCB0008W aka GAL10-ncRNA | New ncRNA antisense to GAL10coordinates 276805..280645 | Houseley et al 2008 PMID:19061643,Pinskaya et al 2009 PMID:19407817, Geisler et al 2012 PMID:22226051 |
| II | YNCB0014W aka TBRT/XUT_2F-154 | New ncRNA antisense to TAT1coordinates 376610..378633 | Awasthi et al 2020 PMID:32081726 |
| III | RE/RE301 | New recombination enhancercoordinates 29108..29809 | Wu and Haber 1996 PMID:8861911 |
| V | YELWdelta27 | New Ty1 LTRcoordinates 449274..449626 | Nene et al 2018 PMID:29320491 |
| V | HPA3/YEL066W | Moved translation start to Met19old coordinates: 26667..27206new coordinates: 26721..27206 | Sampath et al 2013 PMID:23775086 |
| VII | OTO1/YGR227C-A | New ORFcoordinates 949052..949225 Crick | Makanae et al 2015 PMID:25781884 |
| VII | ROK1/YGL171W | Two new uORFscoordinates uORF1: 182286..182407coordinates uORF2: 182291..182329 | Jeon and Kim 2010 PMID:20969870 |
| VIII | YHR052C-B | New ORFcoordinates: 212519..212692 Crick | He et al 2018 PMID:29897761 |
| VIII | YHR054C-B | New ORFcoordinates: 214517..214690 Crick | He et al 2018 PMID:29897761 |
| VIII | SUT169/YNCH0011W | New ncRNAcoordinates 378254..379237 | Xu et al 2009 PMID:19169243,Geisler et al 2012 PMID:22226051, Huber et al 2016 PMID:27292640, Bunina et al 2017 PMID:28977638 |
| X | YJR012C | Moved start to Met76old coordinates: 459795..460418 Cricknew coordinates: 459795..460193 Crick | Sadhu et al 2018 PMID:29632376 |
| X | YJR107C-A | New ORFcoordinates 628457..628693 Crick | Yagoub et al 2015 PMID:26554900, He et al 2018 PMID:29897761 |
| XI | YKL104W-A | New ORFcoordinates 245032..245286 | He et al 2018 PMID:29897761 |
| XII | YLR379W-A | New ORFcoordinates: 877444..877716 | Internal reanalysis of results from Song et al 2015 to find and annotate missing S288C ORFs PMID:25781462 |
| XII | NTS1-2, NTS2-1, NTS2-2 | Change feature_type/SO_term from SO:0001637 rRNA_gene to SO:0000183 non_transcribed_region | |
| XIII | LDO45/YMR147W | Shift stop to be same as LDO16/YMR148W, add intronold coordinates 559199..559870new coordinates 559199..559780, 560156..560812 | Eisenberg-Bord et al 2018 PMID:29187527 |
| XIII | YMR008C-A | New ORFcoordinates 283081..283548 Crick | Internal reanalysis of results from Song et al 2015 to find and annotate missing S288C ORFs PMID:25781462 |
| XIII | YNCM0001W aka PHO84 lncRNA | New ncRNAcoordinates: 23564..26578 | Camblong et al 2007 PMID:18022365 |
| XIV | LTO1/YNL260C | Move start to Met37old coordinates: 156859..157455 Cricknew coordinates: 156859..157347 Crick | Paul et al 2015 PMID:26182403 |
| XVI | YNCP0002W aka GAL4 lncRNA | New ncRNAcoordinates: 79562..82648 | Geisler et al 2012 PMID:22226051 |
New systematic nomenclature system for noncoding RNA genes
| Chr | Systematic_name | Gene_name | Alias | Feature_type | Coordinates | Strand |
|---|---|---|---|---|---|---|
| I | YNCA0001W | HRA1 | ncRNA gene | 99305..99868 | + | |
| I | YNCA0002W | TRN1 | tP(UGG)A, tRNA-Pro | tRNA gene | 139152..139254 | + |
| I | YNCA0003W | SNR18 | snR18 | snoRNA gene | 142367..142468 | + |
| I | YNCA0004W | TGA1 | tA(UGC)A, tRNA-Ala | tRNA gene | 166267..166339 | + |
| I | YNCA0005W | SUP56 | tL(CAA)A, tRNA-Leu | tRNA gene | 181141..181254 | + |
| I | YNCA0006C | tS(AGA)A, tRNA-Ser | tRNA gene | 182522..182603 | – | |
| II | YNCB0001W | tL(UAA)B1, tRNA-Leu | tRNA gene | 9583..9666 | + | |
| II | YNCB0002W | tF(GAA)B, tRNA-Phe | tRNA gene | 36398..36488 | + | |
| II | YNCB0003W | SNR56 | snR56 | snoRNA gene | 88190..88277 | + |
| II | YNCB0004W | tI(AAU)B, tRNA-Ile | tRNA gene | 197494..197567 | + | |
| II | YNCB0005C | tG(GCC)B, tRNA-Gly | tRNA gene | 197629..197699 | – | |
| II | YNCB0006W | tS(AGA)B, tRNA-Ser | tRNA gene | 227075..227156 | + | |
| II | YNCB0007W | tT(AGU)B, tRNA-Thr | tRNA gene | 266378..266450 | + | |
| II | YNCB0008W | GAL10-ncRNA | ncRNA gene | 276805..280645 | + | |
| II | YNCB0009C | SNR161 | snR161 | snoRNA gene | 307185..307345 | – |
| II | YNCB0010W | TLC1 | telomerase RNA gene | 307587..308887 | + | |
| II | YNCB0011C | tV(UAC)B, tRNA-Val | tRNA gene | 326792..326865 | – | |
| II | YNCB0012W | tL(UAA)B2, tRNA-Leu | tRNA gene | 347603..347686 | + | |
| II | YNCB0013W | tQ(UUG)B, tRNA-Gln | tRNA gene | 350827..350898 | + | |
| II | YNCB0014W | TBRT | XUT_2F-154 | ncRNA gene | 376610..378633 | + |
| II | YNCB0015W | tR(UCU)B, tRNA-Arg | tRNA gene | 405878..405949 | + | |
| II | YNCB0016W | tD(GUC)B, tRNA-Asp | tRNA gene | 405960..406031 | + | |
| II | YNCB0017C | tC(GCA)B, tRNA-Cys | tRNA gene | 643007..643078 | – | |
| II | YNCB0018W | tE(UUC)B, tRNA-Glu | tRNA gene | 645167..645238 | + | |
| II | YNCB0019C | LSR1 | snRNA gene | 680688..681862 | – | |
| III | YNCC0001C | tE(UUC)C, tRNA-Glu | tRNA gene | 82462..82533 | – | |
| III | YNCC0002W | SUP53 | tL(CAA)C, tRNA-Leu | tRNA gene | 90859..90972 | + |
| III | YNCC0003C | SNR43 | snR43 | snoRNA gene | 107504..107712 | – |
| III | YNCC0004C | SUF2 | tP(AGG)C, tRNA-Pro | tRNA gene | 123577..123648 | – |
| III | YNCC0005W | tN(GUU)C, tRNA-Asn | tRNA gene | 127716..127789 | + | |
| III | YNCC0006C | SNR33 | snR33 | snoRNA gene | 142364..142546 | – |
| III | YNCC0007C | SUF16 | tG(GCC)C, tRNA-Gly | tRNA gene | 142701..142771 | – |
| III | YNCC0008W | tM(CAU)C, tRNA-Met | tRNA gene | 149920..149991 | + | |
| III | YNCC0009C | tK(CUU)C, tRNA-Lys | tRNA gene | 151284..151356 | – | |
| III | YNCC0010C | tQ(UUG)C, tRNA-Gln | tRNA gene | 168301..168372 | – | |
| III | YNCC0011W | SNR65 | snR65 | snoRNA gene | 177183..177282 | + |
| III | YNCC0012C | SNR189 | snR189 | snoRNA gene | 178610..178798 | – |
| III | YNCC0013W | SUP61 | tS(CGA)C, tRNA-Ser | tRNA gene | 227942..228042 | + |
| III | YNCC0014W | tT(AGU)C, tRNA-Thr | tRNA gene | 295484..295556 | + | |
| IV | YNCD0001W | tG(GCC)D1, tRNA-Gly | tRNA gene | 83548..83618 | + | |
| IV | YNCD0002C | SNR63 | snR63 | snoRNA gene | 323217..323471 | – |
| IV | YNCD0003W | tK(UUU)D, tRNA-Lys | tRNA gene | 359577..359672 | + | |
| IV | YNCD0004W | tA(AGC)D, tRNA-Ala | tRNA gene | 410379..410451 | + | |
| IV | YNCD0005C | tT(AGU)D, tRNA-Thr | tRNA gene | 434264..434336 | – | |
| IV | YNCD0006W | tS(AGA)D1, tRNA-Ser | tRNA gene | 437772..437853 | + | |
| IV | YNCD0007C | tV(UAC)D, tRNA-Val | tRNA gene | 488797..488870 | – | |
| IV | YNCD0008W | tL(UAA)D, tRNA-Leu | tRNA gene | 519743..519826 | + | |
| IV | YNCD0009W | tQ(UUG)D1, tRNA-Gln | tRNA gene | 520972..521043 | + | |
| IV | YNCD0010C | SNR47 | snR47 | snoRNA gene | 541602..541700 | – |
| IV | YNCD0011W | tR(UCU)D, tRNA-Arg | tRNA gene | 568882..568953 | + | |
| IV | YNCD0012W | tD(GUC)D, tRNA-Asp | tRNA gene | 568964..569035 | + | |
| IV | YNCD0013C | tR(ACG)D, tRNA-Arg | tRNA gene | 619969..620041 | – | |
| IV | YNCD0014C | tQ(UUG)D2, tRNA-Gln | tRNA gene | 645153..645224 | – | |
| IV | YNCD0015C | tI(AAU)D, tRNA-Ile | tRNA gene | 668007..668080 | – | |
| IV | YNCD0016C | tQ(UUG)D3, tRNA-Gln | tRNA gene | 802731..802802 | – | |
| IV | YNCD0017W | tI(UAU)D, tRNA-Ile | tRNA gene | 884361..884493 | + | |
| IV | YNCD0018W | SUP2 | tY(GUA)D, tRNA-Tyr | tRNA gene | 946312..946400 | + |
| IV | YNCD0019C | tS(AGA)D2, tRNA-Ser | tRNA gene | 980974..981055 | – | |
| IV | YNCD0020W | tG(GCC)D2, tRNA-Gly | tRNA gene | 992832..992902 | + | |
| IV | YNCD0021C | tE(CUC)D, tRNA-Glu | tRNA gene | 1017207..1017278 | – | |
| IV | YNCD0022C | tV(CAC)D, tRNA-Val | tRNA gene | 1075472..1075544 | – | |
| IV | YNCD0023C | tF(GAA)D, tRNA-Phe | tRNA gene | 1095370..1095461 | – | |
| IV | YNCD0024C | tX(XXX)D | tRNA gene | 1150842..1150941 | – | |
| IV | YNCD0025W | EMT1 | tM(CAU)D, tRNA-Met | tRNA gene | 1175829..1175901 | + |
| IV | YNCD0026W | tK(CUU)D1, tRNA-Lys | tRNA gene | 1201750..1201822 | + | |
| IV | YNCD0027C | SUF3 | tG(CCC)D, tRNA-Gly | tRNA gene | 1257008..1257079 | – |
| IV | YNCD0028W | tS(AGA)D3, tRNA-Ser | tRNA gene | 1305630..1305712 | + | |
| IV | YNCD0029C | tK(CUU)D2, tRNA-Lys | tRNA gene | 1352466..1352538 | – | |
| IV | YNCD0030W | SNR13 | snR13 | snoRNA gene | 1402919..1403042 | + |
| IV | YNCD0031C | tL(CAA)D, tRNA-Leu | tRNA gene | 1461715..1461829 | – | |
| IV | YNCD0032C | SNR84 | snR84 | snoRNA gene | 1492477..1493026 | – |
| V | YNCE0001C | SNR80 | snR80 | snoRNA gene | 52150..52320 | – |
| V | YNCE0002W | SNR67 | snR67 | snoRNA gene | 61352..61433 | + |
| V | YNCE0003W | SNR53 | snR53 | snoRNA gene | 61699..61789 | + |
| V | YNCE0004C | tG(GCC)E, tRNA-Gly | tRNA gene | 61890..61960 | – | |
| V | YNCE0005W | tS(AGA)E, tRNA-Ser | tRNA gene | 86604..86685 | + | |
| V | YNCE0006W | IMT4 | tM(CAU)E, tRNA-Met | tRNA gene | 100133..100204 | + |
| V | YNCE0007C | RPR1 | ncRNA gene | 117667..118035 | – | |
| V | YNCE0008C | tQ(UUG)E2, tRNA-Gln | tRNA gene | 131082..131153 | – | |
| V | YNCE0009C | tK(CUU)E1, tRNA-Lys | tRNA gene | 135425..135497 | – | |
| V | YNCE0010W | tR(UCU)E, tRNA-Arg | tRNA gene | 138666..138737 | + | |
| V | YNCE0011C | SNR14 | snR14 | snRNA gene | 167427..167586 | – |
| V | YNCE0012W | tE(UUC)E1, tRNA-Glu | tRNA gene | 177099..177170 | + | |
| V | YNCE0013W | tH(GUG)E1, tRNA-His | tRNA gene | 207357..207428 | + | |
| V | YNCE0014W | tQ(UUG)E1, tRNA-Gln | tRNA gene | 250286..250357 | + | |
| V | YNCE0015C | SUP19 | tS(UGA)E, tRNA-Ser | tRNA gene | 288443..288524 | – |
| V | YNCE0016C | tA(UGC)E, tRNA-Ala | tRNA gene | 312023..312095 | – | |
| V | YNCE0017W | SRG1 | ncRNA gene | 322212..322762 | + | |
| V | YNCE0018W | tE(UUC)E2, tRNA-Glu | tRNA gene | 354934..355005 | + | |
| V | YNCE0019W | SNR4 | snR4 | snoRNA gene | 424698..424883 | + |
| V | YNCE0020C | SNR52 | snR52 | snoRNA gene | 431129..431220 | – |
| V | YNCE0021C | tH(GUG)E2, tRNA-His | tRNA gene | 434541..434612 | – | |
| V | YNCE0022C | tK(CUU)E2, tRNA-Lys | tRNA gene | 435752..435824 | – | |
| V | YNCE0023W | tV(AAC)E1, tRNA-Val | tRNA gene | 438700..438773 | + | |
| V | YNCE0024W | SCR1 | ncRNA gene | 441987..442508 | + | |
| V | YNCE0025C | tI(AAU)E1, tRNA-Ile | tRNA gene | 443202..443275 | – | |
| V | YNCE0026W | tV(AAC)E2, tRNA-Val | tRNA gene | 469457..469530 | + | |
| V | YNCE0027C | tE(UUC)E3, tRNA-Glu | tRNA gene | 487331..487402 | – | |
| V | YNCE0028C | tR(ACG)E, tRNA-Arg | tRNA gene | 492352..492424 | – | |
| V | YNCE0029C | tI(AAU)E2, tRNA-Ile | tRNA gene | 551285..551358 | – | |
| VI | YNCF0001C | RUF21 | ncRNA gene | 57815..58521 | – | |
| VI | YNCF0002W | SUF9 | tP(UGG)F, tRNA-Pro | tRNA gene | 101376..101478 | + |
| VI | YNCF0003C | RUF20 | ncRNA gene | 131061..131503 | – | |
| VI | YNCF0004C | tN(GUU)F, tRNA-Asn | tRNA gene | 137486..137559 | – | |
| VI | YNCF0005C | tF(GAA)F, tRNA-Phe | tRNA gene | 157916..158007 | – | |
| VI | YNCF0006W | SUF20 | tG(GCC)F1, tRNA-Gly | tRNA gene | 162228..162298 | + |
| VI | YNCF0007W | SUP11 | tY(GUA)F1, tRNA-Tyr | tRNA gene | 167437..167525 | + |
| VI | YNCF0008C | tG(GCC)F2, tRNA-Gly | tRNA gene | 180974..181044 | – | |
| VI | YNCF0009C | tS(GCU)F, tRNA-Ser | tRNA gene | 191513..191613 | – | |
| VI | YNCF0010C | RUF22 | ncRNA gene | 199299..199813 | – | |
| VI | YNCF0011C | tA(AGC)F, tRNA-Ala | tRNA gene | 204924..204996 | – | |
| VI | YNCF0012C | SUP6 | tY(GUA)F2, tRNA-Tyr | tRNA gene | 210619..210707 | – |
| VI | YNCF0013W | RUF23 | ncRNA gene | 221714..221967 | + | |
| VI | YNCF0014C | tK(CUU)F, tRNA-Lys | tRNA gene | 226688..226760 | – | |
| VII | YNCG0001C | RME3 | ncRNA gene | 33109..35013 | – | |
| VII | YNCG0002C | tV(AAC)G3, tRNA-Val | tRNA gene | 73829..73902 | – | |
| VII | YNCG0003C | tH(GUG)G1, tRNA-His | tRNA gene | 110625..110696 | – | |
| VII | YNCG0004W | tK(UUU)G1, tRNA-Lys | tRNA gene | 115488..115583 | + | |
| VII | YNCG0005W | tK(CUU)G1, tRNA-Lys | tRNA gene | 122269..122341 | + | |
| VII | YNCG0006C | RME2 | ncRNA gene | 141898..144120 | – | |
| VII | YNCG0007C | tK(CUU)G2, tRNA-Lys | tRNA gene | 185714..185786 | – | |
| VII | YNCG0008W | tL(CAA)G1, tRNA-Leu | tRNA gene | 205521..205634 | + | |
| VII | YNCG0009C | tW(CCA)G1, tRNA-Trp | tRNA gene | 287350..287455 | – | |
| VII | YNCG0010W | SNR82 | snR82 | snoRNA gene | 316788..317055 | + |
| VII | YNCG0011W | tH(GUG)G2, tRNA-His | tRNA gene | 319781..319852 | + | |
| VII | YNCG0012W | SOE1 | tE(UUC)G1, tRNA-Glu | tRNA gene | 328583..328654 | + |
| VII | YNCG0013W | SNR10 | snR10 | snoRNA gene | 345986..346230 | + |
| VII | YNCG0014C | SNR39 | snR39 | snoRNA gene | 365163..365251 | – |
| VII | YNCG0015C | SNR39B | snR39B | snoRNA gene | 366374..366469 | – |
| VII | YNCG0016C | tE(UUC)G2, tRNA-Glu | tRNA gene | 401527..401598 | – | |
| VII | YNCG0017W | tR(UCU)G1, tRNA-Arg | tRNA gene | 405470..405541 | + | |
| VII | YNCG0018C | tV(AAC)G1, tRNA-Val | tRNA gene | 412294..412367 | – | |
| VII | YNCG0019W | SUP54 | tL(CAA)G2, tRNA-Leu | tRNA gene | 423092..423205 | + |
| VII | YNCG0020C | tF(GAA)G, tRNA-Phe | tRNA gene | 440716..440807 | – | |
| VII | YNCG0021W | tD(GUC)G1, tRNA-Asp | tRNA gene | 531610..531681 | + | |
| VII | YNCG0022W | tE(UUC)G3, tRNA-Glu | tRNA gene | 541850..541921 | + | |
| VII | YNCG0023C | tD(GUC)G2, tRNA-Asp | tRNA gene | 544577..544648 | – | |
| VII | YNCG0024W | SNR46 | snR46 | snoRNA gene | 545370..545566 | + |
| VII | YNCG0025C | tS(AGA)G, tRNA-Ser | tRNA gene | 561662..561743 | – | |
| VII | YNCG0026W | SNR48 | snR48 | snoRNA gene | 609584..609696 | + |
| VII | YNCG0027W | tT(UGU)G1, tRNA-Thr | tRNA gene | 661749..661820 | + | |
| VII | YNCG0028W | tL(GAG)G, tRNA-Leu | tRNA gene | 700675..700756 | + | |
| VII | YNCG0029C | tK(UUU)G2, tRNA-Lys | tRNA gene | 700953..701048 | – | |
| VII | YNCG0030C | tC(GCA)G, tRNA-Cys | tRNA gene | 707108..707179 | – | |
| VII | YNCG0031W | tN(GUU)G, tRNA-Asn | tRNA gene | 731137..731210 | + | |
| VII | YNCG0032W | tR(UCU)G3, tRNA-Arg | tRNA gene | 736340..736411 | + | |
| VII | YNCG0033W | tI(AAU)G, tRNA-Ile | tRNA gene | 739122..739195 | + | |
| VII | YNCG0034W | tA(AGC)G, tRNA-Ala | tRNA gene | 774349..774421 | + | |
| VII | YNCG0035W | SUF4 | tG(UCC)G, tRNA-Gly | tRNA gene | 779616..779687 | + |
| VII | YNCG0036W | tA(UGC)G, tRNA-Ala | tRNA gene | 794417..794489 | + | |
| VII | YNCG0037W | tV(AAC)G2, tRNA-Val | tRNA gene | 823482..823555 | + | |
| VII | YNCG0038C | tR(UCU)G2, tRNA-Arg | tRNA gene | 828723..828794 | – | |
| VII | YNCG0039W | tG(GCC)G1, tRNA-Gly | tRNA gene | 845649..845719 | + | |
| VII | YNCG0040C | tL(CAA)G3, tRNA-Leu | tRNA gene | 857378..857491 | – | |
| VII | YNCG0041W | tK(CUU)G3, tRNA-Lys | tRNA gene | 876394..876466 | + | |
| VII | YNCG0042C | tW(CCA)G2, tRNA-Trp | tRNA gene | 878710..878815 | – | |
| VII | YNCG0043C | tG(GCC)G2, tRNA-Gly | tRNA gene | 930953..931023 | – | |
| VII | YNCG0044C | SNR7-L | snR7-L | snRNA gene | 939459..939672 | – |
| VII | YNCG0045C | SNR7-S | snR7-S | snRNA gene | 939494..939672 | – |
| VII | YNCG0046W | tT(UGU)G2, tRNA-Thr | tRNA gene | 1004216..1004287 | + | |
| VIII | YNCH0001W | tH(GUG)H, tRNA-His | tRNA gene | 62755..62826 | + | |
| VIII | YNCH0002C | tV(AAC)H, tRNA-Val | tRNA gene | 85298..85371 | – | |
| VIII | YNCH0003C | tT(AGU)H, tRNA-Thr | tRNA gene | 116107..116179 | – | |
| VIII | YNCH0004C | tS(AGA)H, tRNA-Ser | tRNA gene | 133026..133107 | – | |
| VIII | YNCH0005W | tQ(UUG)H, tRNA-Gln | tRNA gene | 134321..134392 | + | |
| VIII | YNCH0006C | tA(AGC)H, tRNA-Ala | tRNA gene | 146242..146314 | – | |
| VIII | YNCH0007W | RUF5-1 | ncRNA gene | 212409..213118 | + | |
| VIII | YNCH0008W | RUF5-2 | ncRNA gene | 214407..215116 | + | |
| VIII | YNCH0009C | tF(GAA)H1, tRNA-Phe | tRNA gene | 237848..237939 | – | |
| VIII | YNCH0010C | tF(GAA)H2, tRNA-Phe | tRNA gene | 358478..358569 | – | |
| VIII | YNCH0011W | SUT169 | ncRNA gene | 378254..379237 | + | |
| VIII | YNCH0012W | SNR32 | snR32 | snoRNA gene | 381540..381727 | + |
| VIII | YNCH0013C | SUF8 | tP(UGG)H, tRNA-Pro | tRNA gene | 388893..388995 | – |
| VIII | YNCH0014W | SNR71 | snR71 | snoRNA gene | 411228..411317 | + |
| VIII | YNCH0015W | tT(UGU)H, tRNA-Thr | tRNA gene | 466990..467061 | + | |
| VIII | YNCH0016C | tV(CAC)H, tRNA-Val | tRNA gene | 475706..475778 | – | |
| IX | YNCI0001W | SNR68 | snR68 | snoRNA gene | 97111..97246 | + |
| IX | YNCI0002W | tT(AGU)I1, tRNA-Thr | tRNA gene | 175031..175103 | + | |
| IX | YNCI0003C | tI(AAU)I1, tRNA-Ile | tRNA gene | 183440..183513 | – | |
| IX | YNCI0004W | tE(CUC)I, tRNA-Glu | tRNA gene | 197592..197663 | + | |
| IX | YNCI0005W | tI(AAU)I2, tRNA-Ile | tRNA gene | 210665..210738 | + | |
| IX | YNCI0006W | SUP17 | tS(UGA)I, tRNA-Ser | tRNA gene | 248850..248931 | + |
| IX | YNCI0007C | tK(CUU)I, tRNA-Lys | tRNA gene | 300228..300300 | – | |
| IX | YNCI0008C | tD(GUC)I1, tRNA-Asp | tRNA gene | 324303..324374 | – | |
| IX | YNCI0009W | tT(AGU)I2, tRNA-Thr | tRNA gene | 325748..325820 | + | |
| IX | YNCI0010C | tD(GUC)I2, tRNA-Asp | tRNA gene | 336349..336420 | – | |
| IX | YNCI0011W | tE(UUC)I, tRNA-Glu | tRNA gene | 370417..370488 | + | |
| IX | YNCI0012 | ICR1 | ncRNA gene | 393884..397082 | – | |
| IX | YNCI0013W | PWR1 | ncRNA gene | 395999..396939 | + | |
| X | YNCJ0001C | tT(AGU)J, tRNA-Thr | tRNA gene | 59100..59172 | – | |
| X | YNCJ0002C | tE(UUC)J, tRNA-Glu | tRNA gene | 115939..116010 | – | |
| X | YNCJ0003C | SNR128 | snR128 | snoRNA gene | 139566..139691 | – |
| X | YNCJ0004C | SNR190 | snR190 | snoRNA gene | 139761..139950 | – |
| X | YNCJ0005C | tA(AGC)J, tRNA-Ala | tRNA gene | 197313..197385 | – | |
| X | YNCJ0006W | tD(GUC)J1, tRNA-Asp | tRNA gene | 204735..204806 | + | |
| X | YNCJ0007C | SNR37 | snR37 | snoRNA gene | 228094..228479 | – |
| X | YNCJ0008W | tR(ACG)J, tRNA-Arg | tRNA gene | 233939..234011 | + | |
| X | YNCJ0009C | SNR60 | snR60 | snoRNA gene | 349130..349233 | – |
| X | YNCJ0010W | SUP7 | tY(GUA)J1, tRNA-Tyr | tRNA gene | 354244..354332 | + |
| X | YNCJ0011W | tR(UCU)J1, tRNA-Arg | tRNA gene | 355374..355445 | + | |
| X | YNCJ0012W | tD(GUC)J2, tRNA-Asp | tRNA gene | 355456..355527 | + | |
| X | YNCJ0013C | tD(GUC)J3, tRNA-Asp | tRNA gene | 374424..374495 | – | |
| X | YNCJ0014C | tR(UCU)J2, tRNA-Arg | tRNA gene | 374506..374577 | – | |
| X | YNCJ0015W | tV(AAC)J, tRNA-Val | tRNA gene | 378360..378433 | + | |
| X | YNCJ0016C | EMT5 | tM(CAU)J1, tRNA-Met | tRNA gene | 391043..391115 | – |
| X | YNCJ0017C | tG(GCC)J1, tRNA-Gly | tRNA gene | 396726..396796 | – | |
| X | YNCJ0018W | tK(CUU)J, tRNA-Lys | tRNA gene | 414966..415038 | + | |
| X | YNCJ0019C | tW(CCA)J, tRNA-Trp | tRNA gene | 415931..416036 | – | |
| X | YNCJ0020W | EMT3 | tM(CAU)J2, tRNA-Met | tRNA gene | 422937..423009 | + |
| X | YNCJ0021C | SUP51 | tL(UAA)J, tRNA-Leu | tRNA gene | 424432..424515 | – |
| X | YNCJ0022C | IMT3 | tM(CAU)J3, tRNA-Met | tRNA gene | 517813..517884 | – |
| X | YNCJ0023C | tS(AGA)J, tRNA-Ser | tRNA gene | 524012..524093 | – | |
| X | YNCJ0024W | SUF23 | tG(GCC)J2, tRNA-Gly | tRNA gene | 531828..531898 | + |
| X | YNCJ0025W | HSX1 | tR(CCU)J, tRNA-Arg | tRNA gene | 538555..538626 | + |
| X | YNCJ0026W | tD(GUC)J4, tRNA-Asp | tRNA gene | 541508..541579 | + | |
| X | YNCJ0027C | SUP4 | tY(GUA)J2, tRNA-Tyr | tRNA gene | 542956..543044 | – |
| X | YNCJ0028C | IRT1 | ncRNA gene | 605936..607424 | – | |
| X | YNCJ0029W | tL(UAG)J, tRNA-Leu | tRNA gene | 617919..618019 | + | |
| X | YNCJ0030W | SNR3 | snR3 | snoRNA gene | 663749..663942 | + |
| XI | YNCK0001W | SNR64 | snR64 | snoRNA gene | 38811..38911 | + |
| XI | YNCK0002C | TRT2 | tT(CGU)K, tRNA-Thr | tRNA gene | 46735..46806 | – |
| XI | YNCK0003W | tN(GUU)K, tRNA-Asn | tRNA gene | 74624..74697 | + | |
| XI | YNCK0004W | tL(UAA)K, tRNA-Leu | tRNA gene | 84208..84291 | + | |
| XI | YNCK0005C | tE(UUC)K, tRNA-Glu | tRNA gene | 141018..141089 | – | |
| XI | YNCK0006C | tR(UCU)K, tRNA-Arg | tRNA gene | 162487..162558 | – | |
| XI | YNCK0007W | tK(CUU)K, tRNA-Lys | tRNA gene | 202999..203071 | + | |
| XI | YNCK0008W | tA(AGC)K1, tRNA-Ala | tRNA gene | 219895..219967 | + | |
| XI | YNCK0009W | SNR38 | snR38 | snoRNA gene | 283185..283279 | + |
| XI | YNCK0010W | tW(CCA)K, tRNA-Trp | tRNA gene | 302918..303023 | + | |
| XI | YNCK0011C | tV(AAC)K1, tRNA-Val | tRNA gene | 308144..308217 | – | |
| XI | YNCK0012C | tH(GUG)K, tRNA-His | tRNA gene | 313401..313472 | – | |
| XI | YNCK0013W | SNR69 | snR69 | snoRNA gene | 364776..364876 | + |
| XI | YNCK0014W | tV(AAC)K2, tRNA-Val | tRNA gene | 379680..379753 | + | |
| XI | YNCK0015C | SNR87 | snR87 | snoRNA gene | 431030..431138 | – |
| XI | YNCK0016W | tL(CAA)K, tRNA-Leu | tRNA gene | 458557..458670 | + | |
| XI | YNCK0017W | tR(ACG)K, tRNA-Arg | tRNA gene | 490968..491040 | + | |
| XI | YNCK0018C | tD(GUC)K, tRNA-Asp | tRNA gene | 513332..513403 | – | |
| XI | YNCK0019W | tA(AGC)K2, tRNA-Ala | tRNA gene | 517988..518060 | + | |
| XI | YNCK0020C | SNR42 | snR42 | snoRNA gene | 559016..559366 | – |
| XI | YNCK0021W | tK(UUU)K, tRNA-Lys | tRNA gene | 578965..579060 | + | |
| XII | YNCL0001W | tP(UGG)L, tRNA-Pro | tRNA gene | 92548..92650 | + | |
| XII | YNCL0002C | tS(AGA)L, tRNA-Ser | tRNA gene | 167944..168025 | – | |
| XII | YNCL0003W | SNR30 | snR30 | snoRNA gene | 198784..199389 | + |
| XII | YNCL0004C | tA(UGC)L, tRNA-Ala | tRNA gene | 214883..214955 | – | |
| XII | YNCL0005C | SNR79 | snR79 | snoRNA gene | 348427..348510 | – |
| XII | YNCL0006W | SNR6 | snR6 | snRNA gene | 366235..366346 | + |
| XII | YNCL0007W | tR(ACG)L, tRNA-Arg | tRNA gene | 374355..374427 | + | |
| XII | YNCL0008W | tD(GUC)L1, tRNA-Asp | tRNA gene | 427132..427203 | + | |
| XII | YNCL0009C | tQ(UUG)L, tRNA-Gln | tRNA gene | 448650..448721 | – | |
| XII | YNCL0010C | RDN37-1 | rRNA gene | 451575..458432 | – | |
| XII | YNCL0011C | ETS2-1 | rRNA gene | 451575..451785 | – | |
| XII | YNCL0012C | RDN25-1 | rRNA gene | 451786..455181 | – | |
| XII | YNCL0013C | ITS2-1 | rRNA gene | 455182..455413 | – | |
| XII | YNCL0014C | RDN58-1 | rRNA gene | 455414..455571 | – | |
| XII | YNCL0015C | ITS1-1 | rRNA gene | 455572..455932 | – | |
| XII | YNCL0016C | RDN18-1 | rRNA gene | 455933..457732 | – | |
| XII | YNCL0017C | ETS1-1 | rRNA gene | 457733..458432 | – | |
| XII | YNCL0018W | RDN5-1 | rRNA gene | 459676..459796 | + | |
| XII | YNCL0019C | ETS2-2 | rRNA gene | 460712..460922 | – | |
| XII | YNCL0020C | RDN37-2 | rRNA gene | 460712..467569 | – | |
| XII | YNCL0021C | RDN25-2 | rRNA gene | 460923..464318 | – | |
| XII | YNCL0022C | ITS2-2 | rRNA gene | 464319..464550 | – | |
| XII | YNCL0023C | RDN58-2 | rRNA gene | 464551..464708 | – | |
| XII | YNCL0024C | ITS1-2 | rRNA gene | 464709..465069 | – | |
| XII | YNCL0025C | RDN18-2 | rRNA gene | 465070..466869 | – | |
| XII | YNCL0026C | ETS1-2 | rRNA gene | 466870..467569 | – | |
| XII | YNCL0027W | RDN5-2 | rRNA gene | 468813..468931 | + | |
| XII | YNCL0028W | RDN5-3 | rRNA gene | 472465..472583 | + | |
| XII | YNCL0029W | RDN5-4 | rRNA gene | 482045..482163 | + | |
| XII | YNCL0030W | RDN5-5 | rRNA gene | 485697..485815 | + | |
| XII | YNCL0031W | RDN5-6 | rRNA gene | 489349..489469 | + | |
| XII | YNCL0032C | tL(UAG)L1, tRNA-Leu | tRNA gene | 592519..592619 | – | |
| XII | YNCL0033C | tI(UAU)L, tRNA-Ile | tRNA gene | 605300..605432 | – | |
| XII | YNCL0034W | tL(CAA)L, tRNA-Leu | tRNA gene | 628383..628497 | + | |
| XII | YNCL0035C | tA(AGC)L, tRNA-Ala | tRNA gene | 656934..657006 | – | |
| XII | YNCL0036C | tV(AAC)L, tRNA-Val | tRNA gene | 687859..687932 | – | |
| XII | YNCL0037W | tL(UAG)L2, tRNA-Leu | tRNA gene | 732090..732190 | + | |
| XII | YNCL0038W | tI(AAU)L1, tRNA-Ile | tRNA gene | 734802..734875 | + | |
| XII | YNCL0039W | tX(XXX)L | tRNA gene | 784354..784453 | + | |
| XII | YNCL0040W | tD(GUC)L2, tRNA-Asp | tRNA gene | 793918..793989 | + | |
| XII | YNCL0041C | SNR61 | snR61 | snoRNA gene | 794486..794575 | – |
| XII | YNCL0042C | SNR55 | snR55 | snoRNA gene | 794697..794794 | – |
| XII | YNCL0043C | SNR57 | snR57 | snoRNA gene | 794937..795024 | – |
| XII | YNCL0044W | tE(UUC)L, tRNA-Glu | tRNA gene | 797178..797249 | + | |
| XII | YNCL0045W | TRR4 | tR(CCG)L, tRNA-Arg | tRNA gene | 818609..818680 | + |
| XII | YNCL0046W | SNR44 | snR44 | snoRNA gene | 856710..856920 | + |
| XII | YNCL0047W | tK(UUU)L, tRNA-Lys | tRNA gene | 875376..875471 | + | |
| XII | YNCL0048W | SNR34 | snR34 | snoRNA gene | 899180..899382 | + |
| XII | YNCL0049C | tL(UAA)L, tRNA-Leu | tRNA gene | 962972..963055 | – | |
| XII | YNCL0050C | tN(GUU)L, tRNA-Asn | tRNA gene | 975983..976056 | – | |
| XII | YNCL0051C | tI(AAU)L2, tRNA-Ile | tRNA gene | 1052071..1052144 | – | |
| XIII | YNCM0001W | PHO84 lncRNA | ncRNA gene | 23564..26578 | + | |
| XIII | YNCM0002C | SNR85 | snR85 | snoRNA gene | 67768..67938 | – |
| XIII | YNCM0003W | ZOD1 | ncRNA gene | 91970..92027 | + | |
| XIII | YNCM0004C | tR(UCU)M2, tRNA-Arg | tRNA gene | 131825..131896 | – | |
| XIII | YNCM0005C | SNR54 | snR54 | snoRNA gene | 163535..163620 | – |
| XIII | YNCM0006W | SUP5 | tY(GUA)M1, tRNA-Tyr | tRNA gene | 168795..168883 | + |
| XIII | YNCM0007C | tG(GCC)M, tRNA-Gly | tRNA gene | 183898..183968 | – | |
| XIII | YNCM0008C | SUF7 | tP(UGG)M, tRNA-Pro | tRNA gene | 196068..196170 | – |
| XIII | YNCM0009C | tS(AGA)M, tRNA-Ser | tRNA gene | 259158..259239 | – | |
| XIII | YNCM0010W | tE(UUC)M, tRNA-Glu | tRNA gene | 290801..290872 | + | |
| XIII | YNCM0011W | SNR78 | snR78 | snoRNA gene | 297278..297364 | + |
| XIII | YNCM0012W | SNR77 | snR77 | snoRNA gene | 297506..297593 | + |
| XIII | YNCM0013W | SNR76 | snR76 | snoRNA gene | 297725..297833 | + |
| XIII | YNCM0014W | SNR75 | snR75 | snoRNA gene | 297918..298006 | + |
| XIII | YNCM0015W | SNR74 | snR74 | snoRNA gene | 298138..298225 | + |
| XIII | YNCM0016W | SNR73 | snR73 | snoRNA gene | 298307..298412 | + |
| XIII | YNCM0017W | SNR72 | snR72 | snoRNA gene | 298554..298651 | + |
| XIII | YNCM0018C | tA(AGC)M1, tRNA-Ala | tRNA gene | 321147..321219 | – | |
| XIII | YNCM0019W | tF(GAA)M, tRNA-Phe | tRNA gene | 352280..352370 | + | |
| XIII | YNCM0020W | tH(GUG)M, tRNA-His | tRNA gene | 363064..363135 | + | |
| XIII | YNCM0021C | tV(AAC)M1, tRNA-Val | tRNA gene | 372445..372518 | – | |
| XIII | YNCM0022C | tW(CCA)M, tRNA-Trp | tRNA gene | 379303..379408 | – | |
| XIII | YNCM0023C | tV(AAC)M2, tRNA-Val | tRNA gene | 420588..420661 | – | |
| XIII | YNCM0024W | tD(GUC)M, tRNA-Asp | tRNA gene | 463554..463625 | + | |
| XIII | YNCM0025C | tK(CUU)M, tRNA-Lys | tRNA gene | 480621..480693 | – | |
| XIII | YNCM0026C | SNR24 | snR24 | snoRNA gene | 499984..500072 | – |
| XIII | YNCM0027W | tL(CAA)M, tRNA-Leu | tRNA gene | 504895..505008 | + | |
| XIII | YNCM0028C | EMT4 | tM(CAU)M, tRNA-Met | tRNA gene | 572883..572955 | – |
| XIII | YNCM0029C | tV(AAC)M3, tRNA-Val | tRNA gene | 586636..586709 | – | |
| XIII | YNCM0030W | SNR83 | snR83 | snoRNA gene | 626349..626654 | + |
| XIII | YNCM0031W | SNR11 | snR11 | snoRNA gene | 652275..652532 | + |
| XIII | YNCM0032C | RNA170 | ncRNA gene | 667288..667456 | – | |
| XIII | YNCM0033C | tR(UCU)M1, tRNA-Arg | tRNA gene | 747892..747963 | – | |
| XIII | YNCM0034C | SNR86 | snR86 | snoRNA gene | 762110..763113 | – |
| XIII | YNCM0035C | tA(AGC)M2, tRNA-Ala | tRNA gene | 768369..768441 | – | |
| XIII | YNCM0036C | CDC65 | tQ(CUG)M, tRNA-Gln | tRNA gene | 808246..808317 | – |
| XIII | YNCM0037W | SUP8 | tY(GUA)M2, tRNA-Tyr | tRNA gene | 837928..838016 | + |
| XIV | YNCN0001W | SNR40 | snR40 | snoRNA gene | 89210..89306 | + |
| XIV | YNCN0002C | SUF6 | tG(UCC)N, tRNA-Gly | tRNA gene | 96241..96312 | – |
| XIV | YNCN0003W | tN(GUU)N1, tRNA-Asn | tRNA gene | 102716..102789 | + | |
| XIV | YNCN0004W | tT(AGU)N1, tRNA-Thr | tRNA gene | 104805..104877 | + | |
| XIV | YNCN0005C | SNR19 | snR19 | snRNA gene | 230105..230672 | – |
| XIV | YNCN0006W | tF(GAA)N, tRNA-Phe | tRNA gene | 374869..374959 | + | |
| XIV | YNCN0007W | tL(CAA)N, tRNA-Leu | tRNA gene | 443006..443119 | + | |
| XIV | YNCN0008C | tD(GUC)N, tRNA-Asp | tRNA gene | 519099..519169 | – | |
| XIV | YNCN0009W | tP(UGG)N1, tRNA-Pro | tRNA gene | 547094..547196 | + | |
| XIV | YNCN0010C | tT(AGU)N2, tRNA-Thr | tRNA gene | 560693..560765 | – | |
| XIV | YNCN0011W | tP(UGG)N2, tRNA-Pro | tRNA gene | 568115..568217 | + | |
| XIV | YNCN0012C | tI(AAU)N1, tRNA-Ile | tRNA gene | 569867..569940 | – | |
| XIV | YNCN0013W | NME1 | snoRNA gene | 585587..585926 | + | |
| XIV | YNCN0014W | SNR66 | snR66 | snoRNA gene | 586090..586175 | + |
| XIV | YNCN0015W | tI(AAU)N2, tRNA-Ile | tRNA gene | 602312..602385 | + | |
| XIV | YNCN0016C | SUF10 | tP(AGG)N, tRNA-Pro | tRNA gene | 631846..631917 | – |
| XIV | YNCN0017W | tN(GUU)N2, tRNA-Asn | tRNA gene | 632599..632672 | + | |
| XIV | YNCN0018W | SNR49 | snR49 | snoRNA gene | 716120..716284 | + |
| XIV | YNCN0019C | SNR191 | snR191 | snoRNA gene | 721938..722211 | – |
| XIV | YNCN0020C | tL(UAA)N, tRNA-Leu | tRNA gene | 726134..726217 | – | |
| XV | YNCO0001C | SUF1 | tG(UCC)O, tRNA-Gly | tRNA gene | 110962..111033 | – |
| XV | YNCO0002W | tT(AGU)O1, tRNA-Thr | tRNA gene | 113802..113874 | + | |
| XV | YNCO0003C | SNR58 | snR58 | snoRNA gene | 136088..136183 | – |
| XV | YNCO0004C | tG(GCC)O1, tRNA-Gly | tRNA gene | 226611..226681 | – | |
| XV | YNCO0005W | tN(GUU)O1, tRNA-Asn | tRNA gene | 228331..228404 | + | |
| XV | YNCO0006W | SNR81 | snR81 | snoRNA gene | 234346..234546 | + |
| XV | YNCO0007W | SNR50 | snR50 | snoRNA gene | 259489..259578 | + |
| XV | YNCO0008W | tS(GCU)O, tRNA-Ser | tRNA gene | 274673..274773 | + | |
| XV | YNCO0009W | SUF17 | tG(GCC)O2, tRNA-Gly | tRNA gene | 282164..282234 | + |
| XV | YNCO0010W | SUP3 | tY(GUA)O, tRNA-Tyr | tRNA gene | 288192..288280 | + |
| XV | YNCO0011W | tP(UGG)O1, tRNA-Pro | tRNA gene | 301097..301198 | + | |
| XV | YNCO0012C | tR(ACG)O, tRNA-ARg | tRNA gene | 340299..340371 | – | |
| XV | YNCO0013C | tT(AGU)O2, tRNA-Thr | tRNA gene | 354041..354113 | – | |
| XV | YNCO0014C | SNR9 | snR9 | snoRNA gene | 407948..408134 | – |
| XV | YNCO0015C | SNR62 | snR62 | snoRNA gene | 409765..409864 | – |
| XV | YNCO0016W | tK(UUU)O, tRNA-Lys | tRNA gene | 438643..438738 | + | |
| XV | YNCO0017W | SUF11 | tP(UGG)O2, tRNA-Pro | tRNA gene | 464450..464551 | + |
| XV | YNCO0018W | tN(GUU)O2, tRNA-Asn | tRNA gene | 487439..487512 | + | |
| XV | YNCO0019W | tD(GUC)O, tRNA-Asp | tRNA gene | 571958..572029 | + | |
| XV | YNCO0020C | SUF5 | tG(CCC)O | tRNA gene | 594354..594425 | – |
| XV | YNCO0021C | tV(AAC)O | tRNA gene | 663812..663885 | – | |
| XV | YNCO0022C | SNR36 | snR36 | snoRNA gene | 680685..680866 | – |
| XV | YNCO0023W | IMT1 | tM(CAU)O1, tRNA-Met | tRNA gene | 710201..710272 | + |
| XV | YNCO0024C | SNR35 | snR35 | snoRNA gene | 759326..759529 | – |
| XV | YNCO0025W | SNR17A | snR17a | snoRNA gene | 780107..780596 | + |
| XV | YNCO0026W | SNR8 | snR8 | snoRNA gene | 832332..832521 | + |
| XV | YNCO0027C | SNR31 | snR31 | snoRNA gene | 841958..842182 | – |
| XV | YNCO0028W | SNR5 | snR5 | snoRNA gene | 842403..842606 | + |
| XV | YNCO0029C | tA(UGC)O, tRNA-Ala | tRNA gene | 854187..854259 | – | |
| XV | YNCO0030W | EMT2 | tM(CAU)O2, tRNA-Met | tRNA gene | 976421..976493 | + |
| XV | YNCO0031W | tP(UGG)O3, tRNA-Pro | tRNA gene | 980683..980787 | + | |
| XVI | YNCP0001C | tW(CCA)P, tRNA-Trp | tRNA gene | 56169..56274 | – | |
| XVI | YNCP0002W | GAL4 lncRNA | ncRNA gene | 79562..82648 | + | |
| XVI | YNCP0003W | SNR59 | snR59 | snoRNA gene | 173827..173904 | + |
| XVI | YNCP0004C | tE(UUC)P, tRNA-Glu | tRNA gene | 210192..210263 | – | |
| XVI | YNCP0005C | SNR17B | snR17b | snoRNA gene | 281056..281517 | – |
| XVI | YNCP0006C | IMT2 | tM(CAU)P, tRNA-Met | tRNA gene | 338848..338919 | – |
| XVI | YNCP0007C | tC(GCA)P1, tRNA-Cys | tRNA gene | 435893..435964 | – | |
| XVI | YNCP0008C | tF(GAA)P1, tRNA-Phe | tRNA gene | 560198..560289 | – | |
| XVI | YNCP0009W | tG(GCC)P1, tRNA-Gly | tRNA gene | 572269..572339 | + | |
| XVI | YNCP0010W | tK(CUU)P, tRNA-Lys | tRNA gene | 582062..582134 | + | |
| XVI | YNCP0011C | tF(GAA)P2, tRNA-Phe | tRNA gene | 622540..622631 | – | |
| XVI | YNCP0012W | SUP16 | tS(UGA)P, tRNA-Ser | tRNA gene | 689565..689646 | + |
| XVI | YNCP0013C | SNR51 | snR51 | snoRNA gene | 718700..718806 | – |
| XVI | YNCP0014C | SNR70 | snR70 | snoRNA gene | 718887..719050 | – |
| XVI | YNCP0015C | SNR41 | snR41 | snoRNA gene | 719148..719242 | – |
| XVI | YNCP0016W | tT(UGU)P, tRNA-Thr | tRNA gene | 744284..744355 | + | |
| XVI | YNCP0017C | tK(UUU)P, tRNA-Lys | tRNA gene | 769207..769302 | – | |
| XVI | YNCP0018C | tC(GCA)P2, tRNA-Cys | tRNA gene | 775765..775836 | – | |
| XVI | YNCP0019W | tN(GUU)P, tRNA-Asn | tRNA gene | 810676..810749 | + | |
| XVI | YNCP0020W | tI(AAU)P1, tRNA-Ile | tRNA gene | 819529..819602 | + | |
| XVI | YNCP0021W | SNR45 | snR45 | snoRNA gene | 821732..821903 | + |
| XVI | YNCP0022W | tA(AGC)P, tRNA-Ala | tRNA gene | 856902..856974 | + | |
| XVI | YNCP0023W | tG(GCC)P2, tRNA-Gly | tRNA gene | 860379..860449 | + | |
| XVI | YNCP0024C | tI(AAU)P2, tRNA-Ile | tRNA gene | 880296..880369 | – | |
| Mito | YNCQ0001W | tP(UGG)Q, tRNA-Pro | tRNA gene | 731..802 | + | |
| Mito | YNCQ0002W | 15S_RRNA | Q0020 | rRNA gene | 6546..8194 | + |
| Mito | YNCQ0003W | tW(UCA)Q, tRNA-Trp | tRNA gene | 9374..9447 | + | |
| Mito | YNCQ0004W | tE(UUC)Q, tRNA-Glu | tRNA gene | 35373..35444 | + | |
| Mito | YNCQ0005W | tS(UGA)Q2, tRNA-Ser | tRNA gene | 48201..48290 | + | |
| Mito | YNCQ0006W | 21S_RRNA | Q0158 | rRNA gene | 58009..62447 | + |
| Mito | YNCQ0007W | tT(UGU)Q1, tRNA-Thr | tRNA gene | 63862..63937 | + | |
| Mito | YNCQ0008W | tC(GCA)Q, tRNA-Cys | tRNA gene | 64415..64490 | + | |
| Mito | YNCQ0009W | tH(GUG)Q, tRNA-His | tRNA gene | 64596..64670 | + | |
| Mito | YNCQ0010W | tL(UAA)Q, tRNA-Leu | tRNA gene | 66095..66179 | + | |
| Mito | YNCQ0011W | tQ(UUG)Q, tRNA-Gln | tRNA gene | 66210..66285 | + | |
| Mito | YNCQ0012W | tK(UUU)Q, tRNA-Lys | tRNA gene | 67061..67134 | + | |
| Mito | YNCQ0013W | tR(UCU)Q1, tRNA-Arg | tRNA gene | 67309..67381 | + | |
| Mito | YNCQ0014W | tG(UCC)Q, tRNA-Gly | tRNA gene | 67468..67542 | + | |
| Mito | YNCQ0015W | tD(GUC)Q, tRNA-Asp | tRNA gene | 68322..68396 | + | |
| Mito | YNCQ0016W | tS(GCU)Q1, tRNA-Ser | tRNA gene | 69203..69288 | + | |
| Mito | YNCQ0017W | tR(ACG)Q2, tRNA-Arg | tRNA gene | 69289..69362 | + | |
| Mito | YNCQ0018W | tA(UGC)Q, tRNA-Ala | tRNA gene | 69846..69921 | + | |
| Mito | YNCQ0019W | tI(GAU)Q, tRNA-Ile | tRNA gene | 70162..70237 | + | |
| Mito | YNCQ0020W | tY(GUA)Q, tRNA-Tyr | tRNA gene | 70824..70907 | + | |
| Mito | YNCQ0021W | tN(GUU)Q, tRNA-Asn | tRNA gene | 71433..71503 | + | |
| Mito | YNCQ0022W | tM(CAU)Q1, tRNA-Met | tRNA gene | 72630..72705 | + | |
| Mito | YNCQ0023W | tF(GAA)Q, tRNA-Phe | tRNA gene | 77431..77505 | + | |
| Mito | YNCQ0024C | tT(UAG)Q2, tRNA-Thr | tRNA gene | 78089..78162 | – | |
| Mito | YNCQ0025W | tV(UAC)Q, tRNA-Val | tRNA gene | 78533..78608 | + | |
| Mito | YNCQ0026W | tM(CAU)Q2, tRNA-Met | tRNA gene | 85035..85112 | + | |
| Mito | YNCQ0027W | RPM1 | Q0285 | ncRNA gene | 85295..85777 | + |
Various sequence and annotation files are available on SGD’s Downloads site.
Categories: Data updates
SGD Newsletter, Spring 2021
May 27, 2021
About this newsletter:
This is the Spring 2021 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this newsletter as well as previous newsletters on our Community Wiki.
Contents
- R64.3 Annotation Update
- New Homology Pages
- Functional Complementation Data Available on References Pages
- YeastMine Updates and New Templates
- Textpresso Central Update
- Number of Curated Alleles Continues to Grow
- Alliance of Genome Resources – Disease Associations for model organisms
- Fungal Pathogen Genomics Workshop
R64.3 Annotation Update
SGD curators periodically update the chromosomal annotations of the S. cerevisiae Reference Genome, which is derived from strain S288C.
The R64.3 annotation release, dated 2021-04-21, included various updates and additions:
- 7 new ORFs: OTO1/YGR227C-A, YHR052C-B, YHR054C-B, YJR107C-A, YKL104W-A, YLR379W-A, YMR008C-A
- 5 new ncRNAs: GAL10-ncRNA, TBRT/XUT_2F-154, SUT169, PHO84 lncRNA, GAL4 lncRNA
- 2 new uORFs for ROK1/YGL171W
- 1 new recombination enhancer: RE
- 1 new LTR: YELWdelta27
- 3 ORFs with shifted translation starts: HPA3/YEL066W, YJR012C, LTO1/YNL260C
- 1 ORF with shifted translation stop plus new intron: LDO45/YMR147W
- Changed feature_type (and SO_term) for non-transcribed spacers: NTS1-2, NTS2-1, NTS2-2
- New systematic nomenclature system for entire annotated complement of ncRNAs
Various sequence and annotation files are available on SGD’s Downloads site. You can find more update details and read about the new systematic nomenclature system for noncoding RNA genes on the Details of 2021 Reference Genome Annotation Update R64.3 SGD Wiki page.
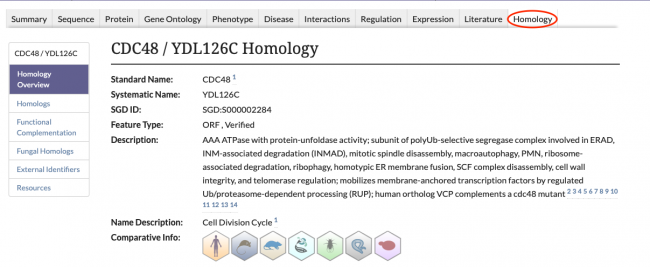
New Homology Pages
SGD is excited to introduce our new Homology Pages! These pages can be accessed by clicking on the Homology tab in the header of SGD gene pages, as seen below.
The information displayed on the Homology Pages is divided into several sections:
- Homologs: Information about known homologs for the gene of interest, such as the species of the homolog, the corresponding Gene ID from the Alliance of Genome Resources, and the name of the homolog.
- Functional Complementation: Data about cross-species functional complementation between yeast and other species, curated by SGD and the Princeton Protein Orthology Database (P-POD).
- Fungal Homologs: Curated homolog information for 24 additional species of fungi. View the species of the fungal homolog, the database source of the entry, and the Gene ID of the homolog from that database.
- External Identifiers: A list of external identifiers for the protein from various database sources.
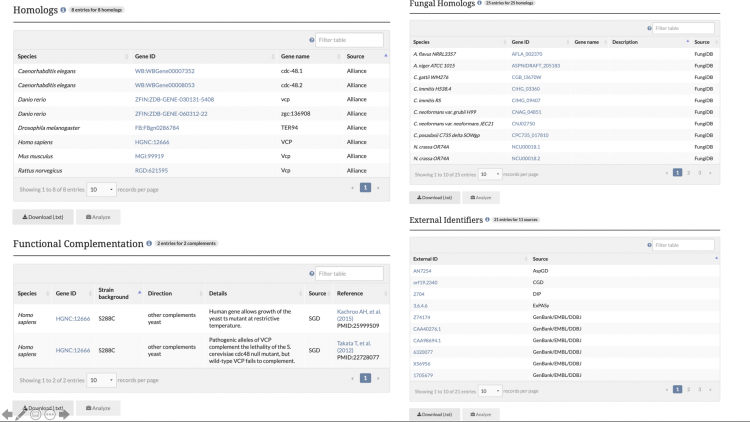
Functional Complementation Data Available on References Pages
Functional Complementation annotations are now viewable on reference pages for which there is curatable functional complementation data. This information describes cross-species functional complementation between yeast and other species, and is curated by SGD and the Princeton Protein Orthology Database (P-POD).
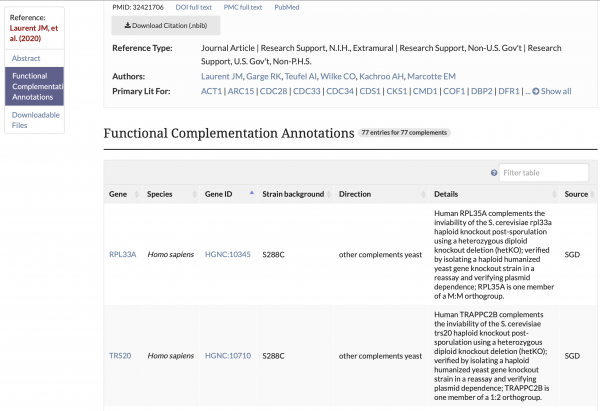
YeastMine Updates and New Templates
SGD has updated the current Gene–>UTRs YeastMine template with newly calculated 5′ and 3′ UTR sequence/coordinates. Additionally, transcript iso-forms for specific genes from the Pelachano et al., 2013 study can be accessed in YeastMine using the new Gene–>Transcripts template. Both templates can be found under the “Templates” section of YeastMine under the “Expression” category.
Textpresso Central Update
Textpresso has recently been updated with a new system, adopting an overhauled user interface and introducing several new features including:
- Search results shown in the context of the full text
- Custom corpus creation
- Customizable annotation interface
- Search terms are highlighted in full-text view
Textpresso Central can also be accessed by clicking on “Full-text Search” under the Literature pull-down menu on the home page of SGD. More information about the changes and types of papers stored in Textpresso can be found in their About Us help section or (from Müller et al., 2018).
Number of Curated Alleles Continues to Grow
SGD now has approximately 13,000 alleles that are either fully or partially curated. To navigate to an allele page, use the search bar to find a specific allele or enter a gene name and select an allele from the autocomplete list. Additionally, these pages can be accessed by clicking on the allele name in a gene’s Phenotype Annotation table. SGD Curators continue to add new alleles or update existing ones as new information becomes available.
You can generate a list of all alleles in our database or find alleles for a specific gene using the Genes –> Alleles template in YeastMine.
Alliance of Genome Resources – Disease Associations for model organisms
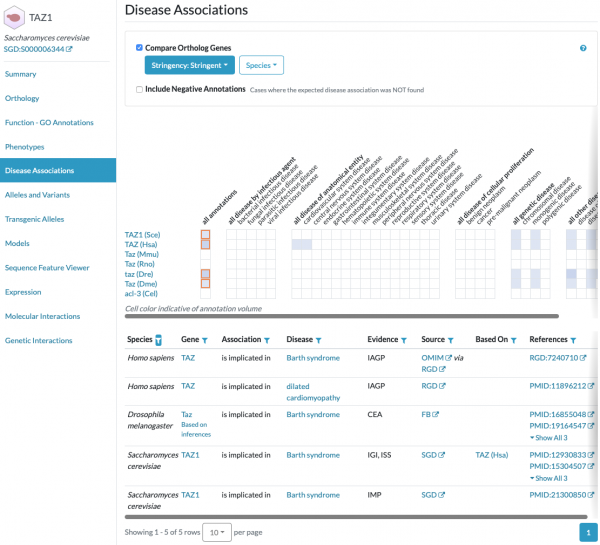
Did you know that you can find human disease associations for yeast genes and their orthologs in other key model organisms at the Alliance of Genome Resources?
SGD is a founding member of the Alliance of Genome Resources, which was established to facilitate the use of diverse model organisms in understanding the genetic and genomic bases of human biology, health, and disease. Gene pages for yeast and other model organisms at the Alliance include a section for Disease Associations, including those for orthologous genes. Human diseases are represented using the Disease Ontology (DO).
Fungal Pathogen Genomics Workshop
From May 10th – 14th, Senior Biocuration Scientist Edith Wong, Senior Biocuration Scientist Rob Nash, Senior Biocuration Scientist Marek Skrzypek, Biocuration Scientist Suzi Aleksander, and Associate Biocuration Scientist Micheal Alexander were instructors for the Virtual Fungal Pathogen Genomics Workshop hosted by Wellcome Connecting Science. Our curators helped attendees learn more about the unique tools hosted on our website and provided them the opportunity to learn about other curation tools from FungiDB, EnsemblFungi, CGD, MycoCosm, and JGI.
We would like to thank the Fungal Pathogen Genomics team for facilitating a successful virtual workshop, and for providing excellent training in web-based data mining resources for all attendees.

Categories: Newsletter
SGD Homology Data Now Available On New Homology Pages
March 25, 2021
SGD is excited to introduce our new Homology Pages! These pages can be accessed by clicking on the Homology tab in the header of SGD gene pages, as seen below.

The information displayed on the Homology Pages is divided into several sections:
- Homologs: Information about known homologs for the gene of interest, such as the species of the homolog, the corresponding Gene ID from the Alliance of Genome Resources, and the name of the homolog.
- Functional Complementation: Data about cross-species functional complementation between yeast and other species, curated by SGD and the Princeton Protein Orthology Database (P-POD).
- Fungal Homologs: Curated homolog information for 24 additional species of fungi. View the species of the fungal homolog, the database source of the entry, and the Gene ID of the homolog from that database.
- External Identifiers: A list of external identifiers for the protein from various database sources.

If you have any questions or feedback regarding our new Homology Pages, please do not hesitate to contact us at any time.
Categories: Data updates, Homologs, New Data, Yeast and Human Disease
BREWMOR Workshop: Preparing Undergraduate Students for Research Experiences
February 03, 2021
BREWMOR: Bridging Research and Education With Model ORganisms (formerly BREW) will be hosting a virtual workshop titled, “Preparing Undergraduate Students for Research Experiences,” on Friday February 19th, 2021 from 4 – 6:30 PM US Eastern time.
After a very successful virtual BREW (Bridging Research and Education Workshop) in July of 2020 as part of the TAGC meeting, a steering committee was formed to coordinate activities of the BREW community. The name of the community was changed to BREWMOR: Bridging Research and Education With Model ORganisms, to include model organisms beyond yeast.
A micro-BREWMOR event that will be held virtually on Friday February 19th, 2021 from 4-6:30 PM US Eastern time. The main purpose of the event is to provide a forum for social interactions and building a community for support and resource sharing. The theme of this micro-BREWMOR will be “Preparing Undergraduate Students for Research Experiences”. The workshop will include a session related to the event’s main theme and opportunities to connect and collaborate with other undergraduate research mentors and teachers in multiple small breakout rooms focused on various topics.
Please register by February 8th at https://forms.gle/fdBCFxYjWuY38tSG6 . Registration is free.
We hope you can join us at the micro-BREWMOR!
https://brewmor.weebly.com/gatherings.html
Categories: Announcements
Apply Now for the 2021 Fungal Pathogen Genomics (Virtual) Course
January 21, 2021

Fungal Pathogen Genomics is an exciting several day long course that provides experimental biologists working on fungal organisms with hands-on experience in genomic-scale data analysis. Through a collaborative teaching effort between the web-based fungal data mining resources FungiDB, EnsemblFungi, PomBase, SGD, CGD, MycoCosm, and JGI, students will learn how to utilize the unique tools provided by each database, develop testable hypotheses, and analyze various ‘omics’ datasets across multiple databases.
Please note: Due to the ongoing Covid-19 pandemic, the 2021 Fungal Pathogen Genomics course will be delivered in a virtual format.
Daily activities will include individual and group training exercises, supplementary lectures on bioinformatics techniques and tools used by various databases, and presentations by distinguished guest speakers covering the following topics:
- Comparative genomics, gene trees, whole-genome alignment
- Identification of orthologs and orthology-based inference
- Genome browsers and gene pages
- RNA-Seq analysis and visualization in VEuPathDB Galaxy
- Variant calling analysis and Ensembl Variant Effect Predictor (VEP) tool
- Development of advanced biologically relevant queries using FungiDB ‘search strategies’ and mining integrated datasets (proteomics, transcriptomics, phenotypes, etc.)
- Genetic interactions, virulence genes, secondary metabolites
- Overview of ontology structure, evidence, available tools, slimming and enrichment
- Introduction to annotation and curation of fungal genomes (e.g. Apollo in EnsemblFungi, FungiDB, and MycoCosm/JGI)
The application deadline for the Fungal Pathogen Genomics workshop to be held May 10-14, 2021 in virtual format is February 18, 2021.
Don’t miss out – apply now!
Categories: Announcements
SGD Newsletter, Fall 2020
December 08, 2020
About this newsletter:
This is the Fall 2020 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this newsletter as well as previous newsletters on our community Wiki.
Contents:
- New SGD Allele Pages
- Interaction Page Updates
- Links on SGD gene pages to homolog gene pages at the Alliance of Genome Resources
- Alliance of Genome Resources at Version 3.2
- Jeremy Thorner Retiring
- YeaZ system for microscopy images
- In Memoriam: Angelika Amon
- Gene Ontology Consortium Meeting
- Happy Holidays from SGD!
New SGD Allele Pages

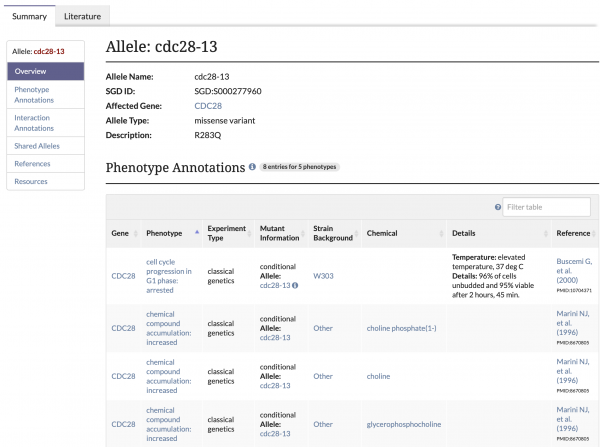
We are pleased to announce that SGD’s brand new Allele Pages are now available on our website. To navigate to an allele page, use the search bar to find a specific allele or enter a gene name and select an allele from the autocomplete list. Additionally, these pages can be accessed by clicking on the allele name in a gene’s Phenotype Annotation table. These pages are still being updated with new information as it becomes available. The type of information that you can find on each allele page includes:
- Allele Overview: General information about the allele, such as its name, the affected gene, the type of allele (e.g. missense), and a description of sequence change and/or domain mutated.
- Phenotype and Interaction Annotations: Phenotype and Genetic Interaction Annotation tables for the allele.
- Shared Alleles: A network diagram depicting shared phenotypes and interactions with other alleles.
If you are interested in viewing all alleles for a specific gene or would like to view a comprehensive list of the alleles that SGD currently has curated, you can use this YeastMine template with your customized parameters.
Interaction Page Updates
SGD has made recent updates to our Gene Interactions Page for improved clarity. Previously, genetic and physical interaction annotations were combined in one table, but now these annotations are recorded in separate annotation tables. The menu in the top left corner can be used to view and navigate to each section of the Interactions page. Additionally, alleles, SGA Scores, and P-values are now included for annotations from the global interactions paper by Costanzo M, et al. (2016) .

Links on SGD gene pages to homolog gene pages at the Alliance of Genome Resources


Previously, the ‘Comparative Info’ section on the Gene Summary page contained a link to the Alliance of Genome Resources if integrated model organism details were available. Now, users will instead find hexagonal buttons representing each model organism (human, mouse, rat, zebrafish, fly, and worm) for which there is homologous gene information at the Alliance of Genome Resources. Clicking on the link will immediately direct the user to the gene page for the selected model organism on the Alliance website.
Alliance of Genome Resources at Version 3.2

The Alliance of Genome Resources , a collaborative effort from SGD and other model organism databases (MOD), released version 3.2 in October. Notable improvements and new features include:
- Gene pages now display transgenic allele data under the ‘Transgenic Allele’ section.
- Allele and Variant sections on Gene pages have improved formatting and are now downloadable.
- “NOT” disease annotations are shown in Disease, Alleles and Variants, and Models sections when an anticipated disease annotation was not found.
- Enhanced variant data representation on Allele pages.
- High-throughput profiling experiment metadata can be searched and will display in the search results as HTP Dataset Index.
- The Alliance User Community discussion forum has been released.
Jeremy Thorner Retiring

Jeremy Thorner, a prolific yeast researcher and outstanding mentor, has announced that his lab is closing. His lab provided the following notice:
“After 47 years at the University of California, Berkeley, the laboratory of Professor Jeremy Thorner will be closing permanently, as of 30 June 2021. After that date, there will be no way to distribute any strains, plasmids, enzymes, or antibodies generated during the course of the studies on Saccharomyces cerevisiae conducted by the Thorner laboratory over those many years.”
YeaZ system for microscopy images

YeaZ is a system for efficiently and accurately segmenting microscopy images of yeast cells. It contains a convolutional neural network, with an underlying training set of high-qual|ty segmented yeast images, as well as a graphical user interface and a web application to employ, test, and expand the system. The system contains a Python based application with graphical user interface available on GitHub , as well as standalone apps for both Windows and Mac based computers, and training sets. Additional information is available in the accompanying Nature Communications paper by Dietler et al., 2020 .
YeaZ was created at École polytechnique fédérale de Lausanne (EPFL), Lausanne, Switzerland. Please contact Sahand Jamal Rahi with questions, or ideas for improvements.
In Memoriam: Angelika Amon

It was with great sadness that we learned that Angelika Amon, cell biologist and Professor of Biology at MIT and a member of the Koch Institute for Integrative Cancer Research, passed away on October 29, 2020. Angelika was widely known for her novel contributions to the field of cell biology and proliferation, with her research focusing on aneuploidy and the consequences of chromosome mis-segregation. At the beginning of Angelika’s career, her work on yeast genetics led to the discovery that cyclins must break down completely before cells progress from mitosis to G1.
Her research has helped shape the current understanding of cell division, and her passion for genetics will live on through her students and colleagues who continue her work. Angelika was a treasured part of the genetics community and will be missed dearly.
The Genetics Society of America has featured a tribute piece in remembrance of Angelika Amon , written by her friend and colleague, Orna Cohen-Fix.
Gene Ontology Consortium Meeting

From October 6th-8th, PI Mike Cherry, Senior Biocuration Scientist Edith Wong, Senior Biocuration Scientist Rob Nash, Senior Biocuration Scientist Marek Skrzypek, Biocuration Scientist Suzi Aleksander, and Associate Biocuration Scientist Micheal Alexander attended the Gene Ontology Consortium meeting. Suzi presented on SGD’s GO display and GO tools, and curators learned more about how other databases utilize GO. SGD Curators also participated in meaningful discussions about improving existing GO resources while also helping with the planning of new GO projects.
We would like to thank The Gene Ontology for facilitating this successful change to a virtual conference and holding an accessible, well received event for the entire model organism community!
Happy Holidays from SGD!
We would like to take this opportunity to recognize that 2020 has brought many changes and challenges for everyone. Our thoughts go out to all those who have been impacted by the unprecedented events of this year.
We wish you and your family, friends, and lab mates the best during the upcoming holidays. Stanford University will be closed for three weeks starting on December 14 and will reopen on January 4th, 2021 . Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year.
Categories: Newsletter
Interactions Page Updates
November 06, 2020
SGD has made recent updates to our Gene Interactions Page for improved clarity. Previously, genetic and physical interaction annotations were combined in one table, but now these annotations are recorded in separate annotation tables. The menu in the top left corner can be used to view and navigate to each section of the Interactions page.
Additionally, alleles, SGA Scores, and P-values are now included for annotations from the global interactions paper by Costanzo M, et al. (2016).

Please be sure to watch our Interactions Page Updates tutorial video for a quick walk-through of the changes:
If you have any questions or feedback about the updates to our Interactions Pages, please do not hesitate to contact us at any time.
Categories: Announcements, Data updates
Explore SGD Allele Data Using New Allele Pages
October 29, 2020
We are pleased to announce that SGD’s brand new Allele Pages are now available on our website. To navigate to an allele page, use the search bar to find a specific allele or enter a gene name and select an allele from the autocomplete list. Additionally, these pages can be accessed by clicking on the allele name in a gene’s Phenotype Annotation table. These pages are still being updated with new information as it becomes available.
The type of information that you can find on each allele page includes:
- Allele Overview: General information about the allele, such as its name, the affected gene, the type of allele (e.g. missense), and a description of sequence change and/or domain mutated.
- Phenotype and Interaction Annotations: Phenotype and Genetic Interaction Annotation tables for the allele.
- Shared Alleles: A network diagram depicting shared phenotypes and interactions with other alleles.
Start browsing the thousands of alleles we now have yourself, or check out our Allele Pages video for a more detailed walk-through of what you can expect to find on each allele page:
If you are interested in viewing all alleles for a specific gene or would like to view a comprehensive list of the alleles that SGD currently has curated, you can use this YeastMine template with your customized parameters.

If you have any questions or feedback about the new Allele pages and data, please don’t hesitate to contact us at any time.
Categories: Website changes
Announcement from Jeremy Thorner
October 27, 2020
NOTICE: After 47 years at the University of California, Berkeley, the laboratory of Professor Jeremy Thorner will be closing permanently, as of 30 June 2021. After that date, there will be no way to distribute any strains, plasmids, enzymes, or antibodies generated during the course of the studies on Saccharomyces cerevisiae conducted by the Thorner laboratory over those many years.
Categories: Announcements